Detection and genetic analysis of zoonotic hepatitis E virus, rotavirus, and sapovirus in pigs
Article information
Abstract
The zoonotic transmission of viral diseases to humans is a serious public health concern. Pigs are frequently a major reservoir for several zoonotic viral diseases. Therefore, periodic surveillance is needed to determine the infection rates of zoonotic diseases in domestic pigs. Hepatitis E virus (HEV), rotavirus, sapovirus (SaV), and norovirus (NoV) are potential zoonotic viruses. In this study, 296 fecal samples were collected from weaned piglets and growing pigs in 13 swine farms, and the viral RNA was extracted. Partial viral genomes were amplified by reverse transcription-polymerase chain reaction (PCR) or nested-PCR using virus-specific primer sets under different PCR conditions. HEV-3, rotavirus A, and SaV genogoup 3 were detected from 11.5, 2.7, and 3.0% of the samples, respectively. On the other hand, NoV was not detected in any of the samples. Genetic analysis indicated that the nucleotide sequences of swine HEV-3 and rotavirus A detected in this study were closely related to those of human isolates. However, swine SaV was distant from the human strains. These results suggest that HEV-3 and rotavirus A can be transmitted from pigs to humans. Therefore, strict preventive measures should be implemented by workers in the swine industry to prevent infections with HEV-3 and rotavirus A excreted from pigs.
Introduction
Zoonotic transmissions of viral diseases from pigs to humans have been increasing [1,2]. Because pigs are a major reservoir for zoonotic viruses, continuous surveillance is important to determine the infection status of zoonotic viral diseases in pig populations [3-5]. People who frequently come in contact with animals, such as farmers, veterinarians, and slaughters, are highly exposed to zoonotic diseases. The increase in pork consumption by people would be another risk factor for the transmission of zoonotic viral diseases to humans [6,7].
Hepatitis E virus (HEV), rotavirus, norovirus (NoV), and sapovirus (SaV) are considered potential zoonotic agents that can be transmitted to humans from pigs [8-11]. HEV is a non-enveloped, positive-sense single-stranded RNA virus. The genome size is approximately 7.5 kb. HEV is in the family Hepeviridae that has been classified into eight genotypes in the species Orthohepavirus A [12]. The rotavirus in the family of Reoviridae is non-enveloped, double-stranded RNA virus with 0.7-3.3 kb 11 segmented genomes [13]. Rotavirus can be classified into nine spices: A, B, C, D, E, F, G, H, and I. Among them, group A rotavirus is the major cause of viral diarrhea in humans and several animal species. SaV and NoV are non-enveloped, positive-sense single-stranded RNA viruses in the family Caliciviridae [13,14]. NoV can be classified into at least seven genogroups (GI-GVII) [15]. SaV has been classified into at least 15 genogroups (GI-GXV) [16].
HEV infections generally cause acute self-limiting hepatitis, with approximately 2% mortality. On the other hand, acute liver failures and high mortality, up to 20-30%, have been reported in pregnant women in some developing countries, such as India [17,18]. More than 20 million individuals are infected with HEV per year worldwide. In 2010, HEV infections caused 60,000 deaths worldwide [19]. Pigs infected with HEV show no clinical signs of hepatitis, but humans who eat raw or undercooked pork can develop acute hepatitis [20,21].
Rotaviruses are the major agents of viral diarrhea in young children, newborn piglets, and calves [3,13]. Approximately 215,000 children under five years of age die each year by gastroenteritis caused by rotavirus infections, according to the reports from the World Health Organization [22]. NoV infections are responsible for 47-97 and 5-36% of outbreaks and sporadic cases of acute gastroenteritis worldwide, respectively [23]. SaV can cause acute gastroenteritis in humans of all ages. On the other hand, SaV infections induce milder diarrhea than NoV infections in people [24]. Porcine strains of NoV and SaV that are closely related to human strains appear to cause mild diarrhea in pigs [4,25]. Therefore, pigs may play an important role in the transmission of NoV and SaV to humans [11].
In this study, the partial genomic sequences of HEV, rotavirus, and SaV were detected from fecal samples of pigs, and their genes were analyzed. Considerable portions of pigs were infected with the aforementioned viral diseases.
Materials and Methods
Fecal samples and RNA extraction
A total of 296 normal fecal samples were collected from 13 swine farms selected randomly in South Korea. Among the fecal samples, 142 and 154 samples were obtained from weaned piglets (5-8 weeks old) and growing pigs (9-16 weeks old), respectively. A fecal suspension of each sample was prepared for the extraction of viral RNA. The samples were diluted 1:10 (w/v) with phosphate-buffered saline (PBS) and centrifuged at 3,000 × g for 30 min. The supernatants were collected and stored at -70°C. The viral RNA was extracted from 150 μl of the supernatant using a Patho Genespin DNA/RNA kit (iNtRON Biotechnology, Korea) according to the manufacturer’s instructions. The purity and concentration of the viral RNA were determined using a NanoDrop ND-1000® (Thermo Scientific, USA). The viral RNA extracted from each fecal sample was stored at -70°C.
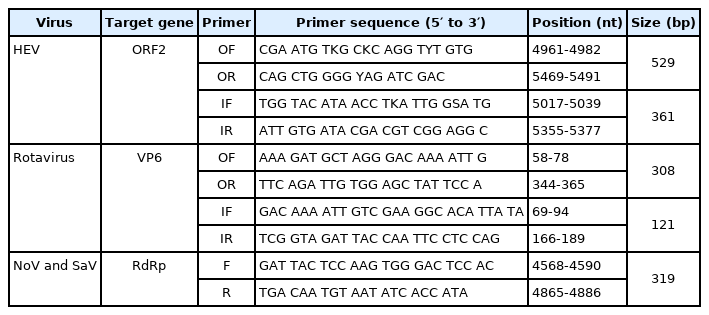
Primer, reverse transcription polymerase chain reaction (RT-PCR), and nested PCR
RT-PCR and PCR kits (Invitrogen, USA) were used for the synthesis of cDNA and RT-PCR process according to the manufacturer’s instructions. Table 1 lists the virus-specific primer sets, their sequences, and the sizes of the PCR products [8,26,27]. To detect HEV and rotavirus, the viral RNA was reverse transcribed to single-stranded cDNA at 45°C for 30 min with the HEV-specific outer reverse (OR) primer and rotavirus-specific OR primer. The first PCR was conducted with the HEV-specific outer forward (OF) and OR primer sets under the following thermal cycling conditions: 35 cycles of denaturation at 94°C for 30 sec, annealing at 48°C for 30 sec, and extension at 72°C for 1 min. Nested-PCR was conducted using the HEV-specific inner forward (IF) and inner reverse (IR) primer set under the following thermal cycling conditions: 35 cycles of denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, and extension at 72°C for 1 min. The first PCR for the detection of rotavirus was conducted using the rotavirus-specific OF and OR primer set under the following conditions: 35 cycles of denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, and extension at 72°C for 1 min. Nested-PCR was conducted with the rotavirus-specific IF and IR primer set under the following conditions: 35 cycles of denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, and extension at 72°C for 1 min. NoV and SaV were detected by one-step RT-PCR under the following conditions: reverse transcription of viral RNA to single-stranded cDNA with the reverse primer specific to NoV and SaV at 45°C for 30 min, and 40 cycles of denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, and extension at 72°C for 1 min with the NoV and SaV-specific forward and reverse primer sets.
Cloning and DNA sequencing
The PCR products were separated by electrophoresis in a 2% agarose gel and visualized under ultraviolet light to identify the expected sizes of the amplified viral genes. Selected PCR products were purified using a MEGA quick-spin total fragment DNA purification kit (iNtRON Biotechnology, Korea). The purified PCR products were ligated into the TA cloning vector, which was transformed to HIT Competent Cells-DH5α (RBC Corporation, Taiwan). The plasmid DNA extracted from the bacterial clone was used to confirm the viral DNA sequences (Cosmogenetech, Korea).
Phylogenetic analysis
Each viral partial genomic sequence obtained in this study was aligned with the representative viral genes deposited in GenBank using BioEdit 7.2.5 software. Phylogenetic analysis was conducted using the confirmed viral sequences obtained in this study using MEGA software (ver.7; Kumar, Stecher, and Tamura 2015) according to the maximum likelihood method. Boot trapping was performed with 500 replicates.
Results
Prevalence rates of HEV, rotavirus, and SaV
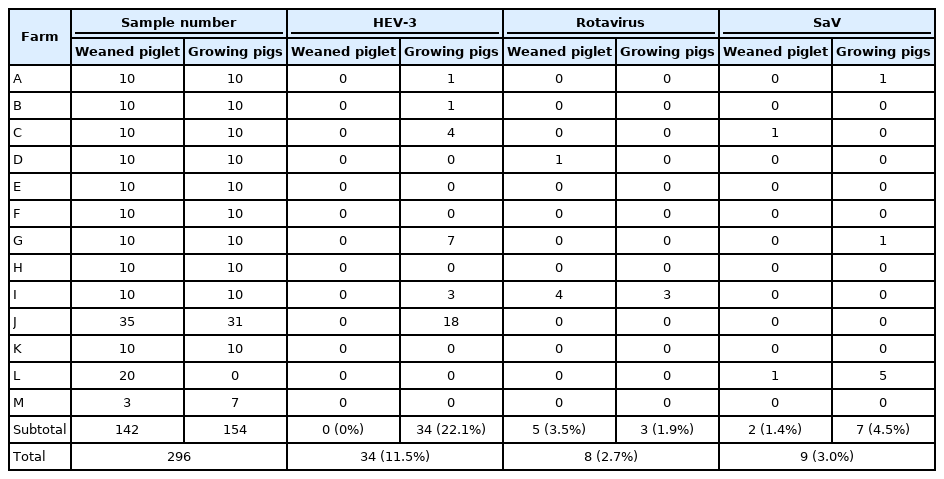
To determine the prevalence of the HEV, rotavirus, SaV, and NoV in pigs, 296 fecal samples were collected from 13 farms. The samples were screened for the presence of viral genes by RT-PCR and nested PCR using each virus-specific primer set (Table 1). HEV-3 was detected in 34/154 (22.1%) of growing pigs but none (0/142) of the weaned piglets showing a prevalence rate of 11.5% (Table 2). Rotavirus A was detected in 5/142 (3.5%) and 3/154 (1.9%) of weaned and growing pigs, respectively, showing a total prevalence rate of 2.7% (Table 2). SaV was detected in 2/142 (1.4%) of weaned piglets and 7/154 (4.5%) of growing pigs, showing a total prevalence rate of 3.0% (Table 2). NoV was not detected in any of the fecal samples.
Phylogenetic analysis of viral genes
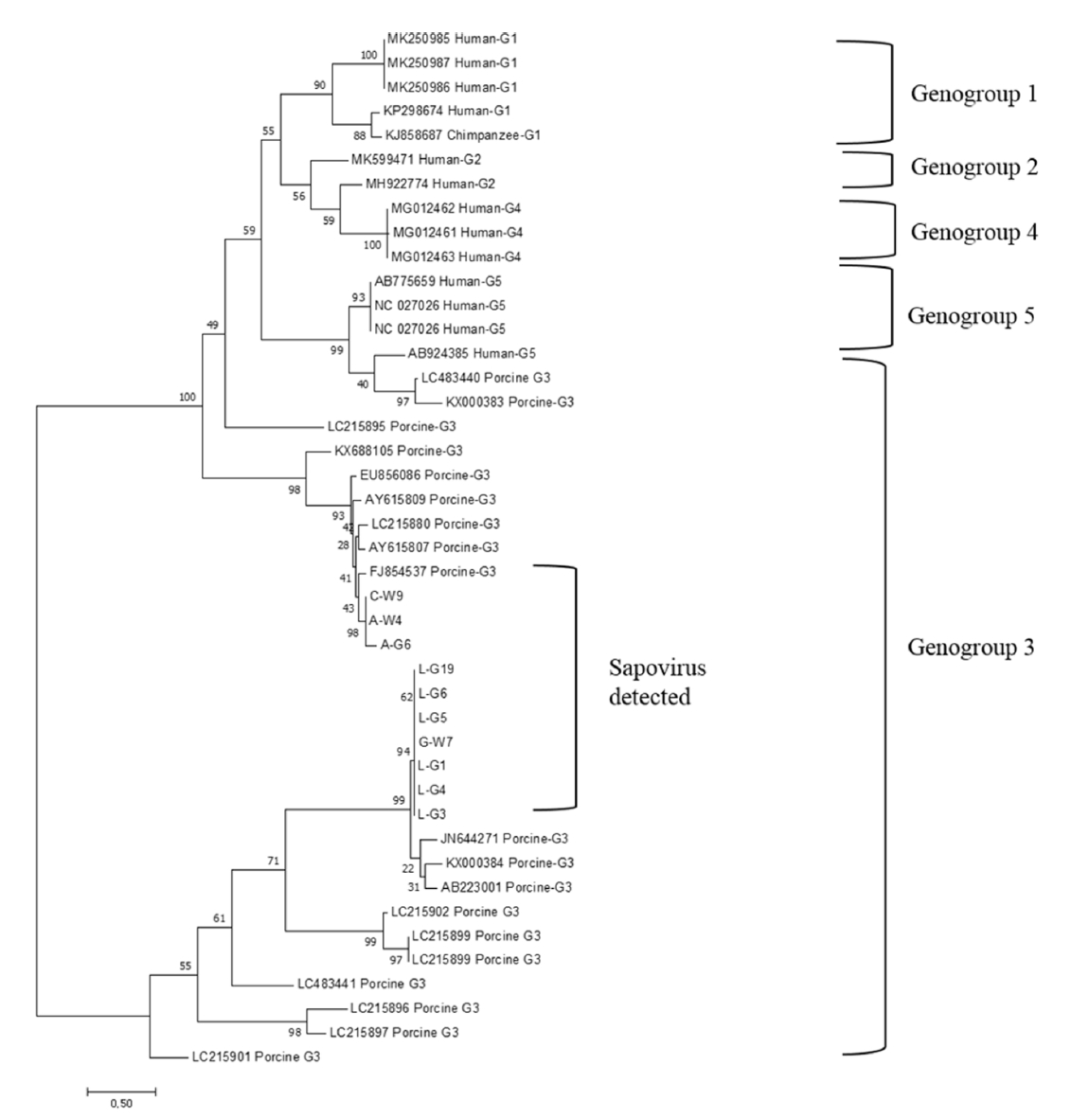
All the HEV strains isolated in this study were analyzed phylogenetically with the representative strains of HEV-1, -2, -3, and -4. They were all classified into HEV-3 and were closely related to swine HEV-3, which was previously reported from Korea (GenBank accession numbers: FJ426403 and FJ426404) (Fig. 1). Their nucleotide sequences showed 96% nucleotide identity with the two representative HEV strains (data not shown). All the rotavirus strains isolated in this study were classified into the rotavirus A group (Fig. 2). They were closely related to four human strains of rotavirus A (GenBank accession numbers: FJ947400, KC580124, KJ659479, and LC485135). The strains had 93.4-96% nucleotide identity with the human rotavirus strains (data not shown). All the SaV strains isolated in this study were classified into genogroup 3 and were closely related to four porcine SaV strains (GenBank accession numbers: JN644271, KX000384, AB223001, and FJ854537) (Fig. 3). Their nucleotide sequence shared 84-91% identities with those of the porcine SaV strains (data not shown).
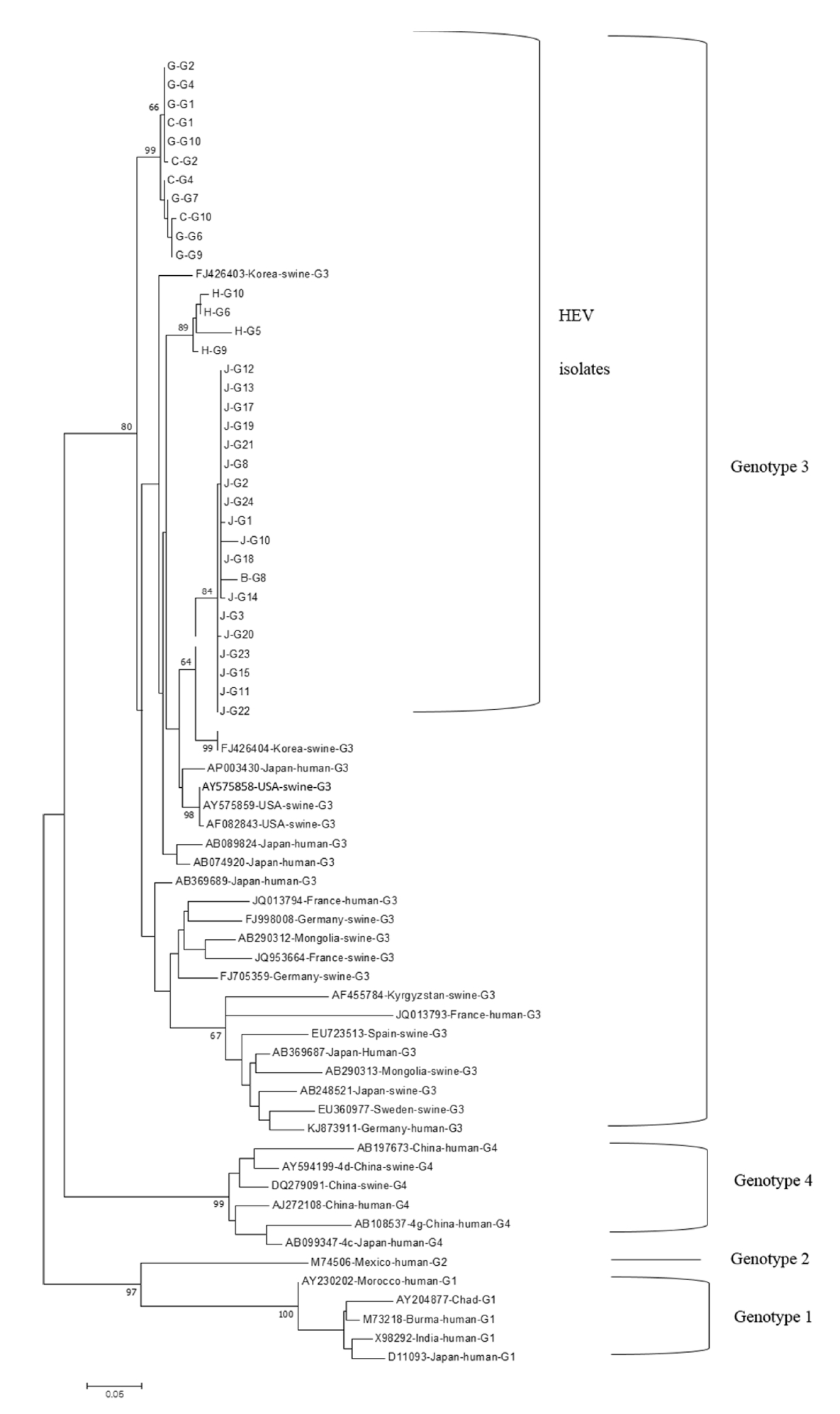
Phylogenetic analysis of HEV nucleotide sequences obtained from this study with other HEV strains. The total 360-bp nucleotide sequences positioned at the junction of ORF1 and ORF2 were analyzed with the representative HEV-1, HEV-2, HEV-3, and HEV-4 strains. All 34 HEV-3 strains were detected from growing pigs. The sample identities were named with the farm names (A-M), group names (G-growing pigs, W-weaned piglets), and sample number. HEV, hepatitis E virus;ORF, open reading frame.

Phylogenetic analysis of rotavirus genomic sequence. The total 121-bp nucleotide sequences of VP6 gene obtained from eight rotaviruses were analyzed with those of rotaviruses representing groups A, B, C, D, F, G, H, and I.
Discussion
HEV was first found in the early 1980s in New Delhi, India. Infections with HEV-1 and HEV-2 only occur in humans in developing countries. On the other hand, HEV-3, HEV4, and HEV-7 cause infections in several animal species and humans. Therefore, the zoonotic transmission of these HEVs is a major public health concern, mainly in developed countries. In Korea, zoonotic transmission cases of HEV have been reported [28]. In a recent study, HEV-3 and HEV-4 were detected in 20.3% of growing pigs in Korea [29]. In this study, HEV-3 was found in 22.1% of the fecal samples of growing pigs. The results indicated similar prevalence rates of HEV in growing pigs in two different studies carried out in Korea. On the other hand, only HEV-3 was found in this study without the detection of HEV-4. HEV-4 was not detected because of the different regions from which swine fecal samples were collected [29]. All sequences of HEV-3 detected in this study were similar to the swine HEV3 strains reported previously in Korea [5]. This indicates that the HEV-3 strains detected in this study might have originated from old HEV-3 strains prevailing in pigs in Korea. On the other hand, their sequences were also closely related to those of human HEV strains in Japan. The close genetic relationships of swine HEV-3 determined in this study with human HEV-3 strains suggested the possible transmission of swine HEV to humans.
Rotavirus causes diarrhea, mainly in young children and several animal species. Rotavirus infections of piglets and calves cause severe economic losses in the animal husbandry industry [9,30]. In particular, group A rotavirus is recognized as a zoonotic virus and the major causative agent infecting young children and domestic animals [13,31]. The transmissions of group A rotavirus from animal species to humans have been reported [10,32]. In addition, piglets can be infected experimentally with human rotavirus. In this study, group A rotavirus was detected in 3.5 and 1.9% of fecal samples of weaned piglets and growing pigs, respectively. The genomic sequences of rotavirus A determined in this study were close to those of the human rotavirus strains [33,34]. These results suggest that some of normal fecal samples excreted from healthy-looking pigs contain rotavirus A. In another study, rotavirus A could be found in 38.3% of diarrheic fecal samples of pigs [35]. Therefore, zoonotic rotavirus A appeared to be secreted mainly from pigs with diarrhea but also some healthy pigs.
NoV and SaV are important enteric viruses that cause acute gastroenteritis in humans and possibly in pigs. They usually cause food- or water-borne illnesses in humans. On the other hand, it has not been clearly defined whether NoV can cause severe diarrhea in pigs. No animal NoV was detected in human stool samples [11]. Nevertheless, the zoonotic transmission of animal NoV would be possible because of the close genetic relationship between animal NoV and human NoV [25,36]. Although SaV infects infants and young children, they induce milder diarrhea than NoV [24]. Hence, SaV might cause mild gastroenteritis in pigs. Porcine SaV also has close genetic relationships to human SaV [37]. In this study, NoV was not found in any fecal sample of pigs. On the other hand, SaV was detected in 1.4 and 4.5% of weaned piglets and growing pigs, respectively. Previous studies reported prevalence rates of 0.5-1.9% for NoV and 6.5-11.2% for SaV in pigs in Korea [38,39]. SaV determined in this study was classified into the genogroup 3 porcine strain of SaV [16,40]. Their genomic sequences were closely related to porcine SaV reported from Denmark [40]. These results suggest that the SaV strains found in Korea might have been introduced from a foreign country.
The main purpose of this study was to determine the prevalence rates of four zoonotic viruses: HEV, rotavirus, NoV, and SaV. Therefore, this study focused on the detection of the highly conserved genomic sequences of the viruses instead of determining their full genomic sequences. A future study will try to determine the full genomic sequences of these viruses.
In conclusion, this study showed that HEV-3 and rotavirus detected from pigs have closely related genomic sequences to human viruses. On the other hand, SaV determined in this study was close to the porcine strains found in a foreign country. Therefore, the possible zoonotic transmission of HEV and rotavirus would be higher than SaV. Farmworkers, veterinarians, and other people who frequently contact pigs need to take personal measures to prevent the zoonotic transmission of these viruses.
Notes
The authors declare no conflict of interest.
Acknowledgements
This study was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C 1836).