|
 |
| Korean J Vet Res > Volume 63(2); 2023 > Article |
|
Abstract
Acknowledgments
Fig. 1.
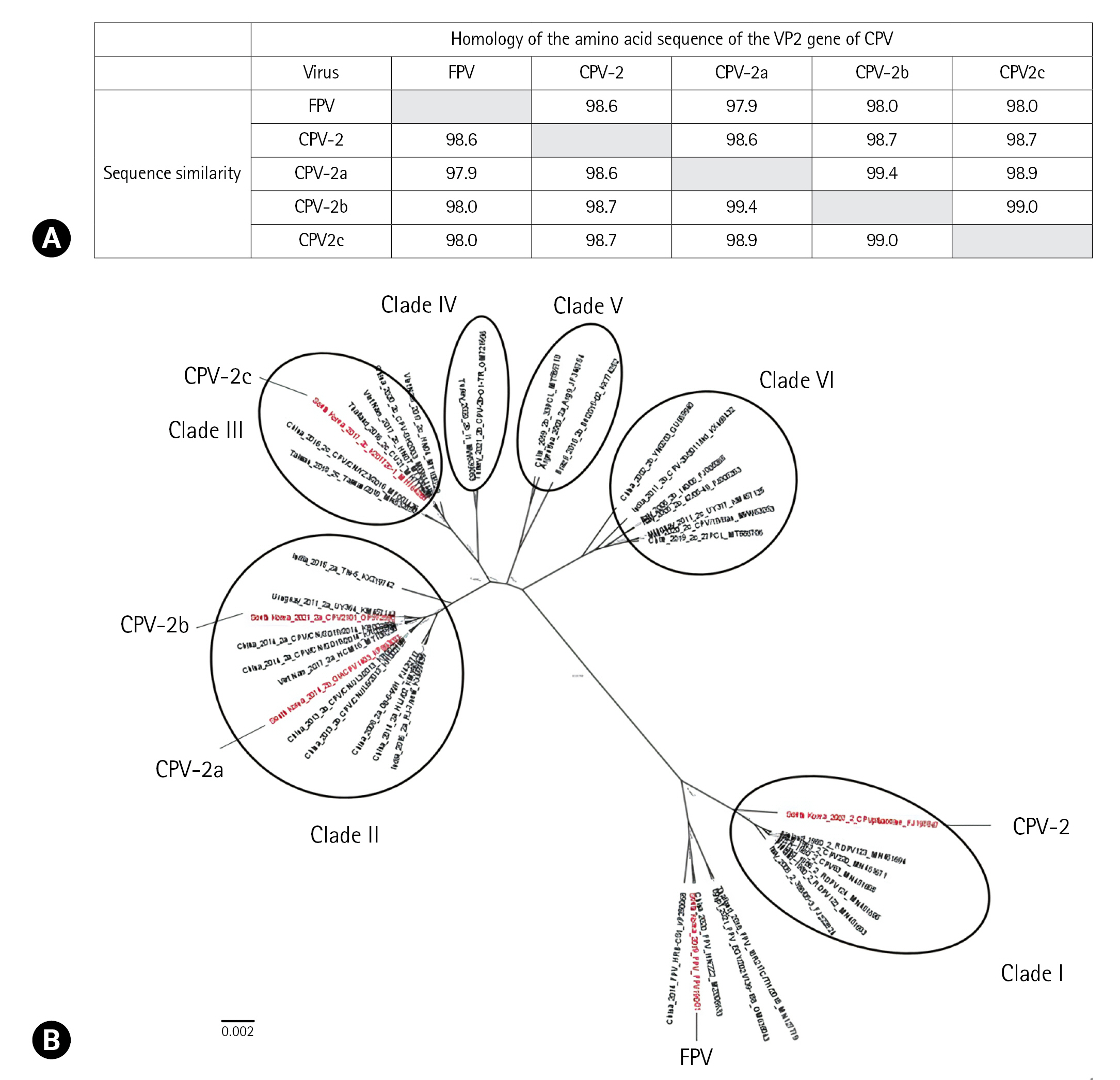
Table 1.
Strength score of reactions as judged by eye: 3, strong; 2, medium; 1, weak; 0.5, very weak; 0, negative.
CAV-1, canine adenovirus type 1; CAV-2, canine adenovirus type 2; CRCoV, canine respiratory coronavirus; CDV, canine distempervirus; PIV-5, parainfluenza virus type 5; CIV, canine influenza virus (H3N2); RABV, rabies virus; RoV, rotavirus.
Table 2.
| Company |
Genotype |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
CPV-2 |
CPV-2a |
CPV-2b |
CPV-2c |
FPV |
|||||||||||
| 6* | 5 | 4 | 6 | 5 | 4 | 6 | 5 | 4 | 6 | 5 | 4 | 6 | 5 | 4 | |
| A | 0 | 0 | 0 | 3 | 2 | 0.5 | 2 | 1 | 0.5 | 2 | 1 | 0.5 | 0 | 0 | 0 |
| B | 3 | 2 | 0.5 | 3 | 3 | 1 | 3 | 3 | 1 | 3 | 3 | 1 | 2 | 1 | 0 |
| C | 0 | 0 | 0 | 3 | 2 | 0.5 | 2 | 1 | 0.5 | 2 | 1 | 0 | 0 | 0 | 0 |
| D | 2 | 1 | 0 | 3 | 2 | 0.5 | 2 | 1 | 0.5 | 2 | 1 | 0.5 | 1 | 0.5 | 0 |
| E | 3 | 2 | 0.5 | 3 | 2 | 0.5 | 3 | 2 | 0.5 | 3 | 2 | 0.5 | 2 | 0.5 | 0 |
| F | 3 | 2 | 0.5 | 2 | 1 | 0 | 0 | 0 | 0 | 3 | 2 | 0.5 | 2 | 0.5 | 0 |
| Mean ± SD | 1.8 ± 1.3 | 1.2 ± 0.9 | 0.25 ± 0.3 | 2.8 ± 0.4 | 2 ± 0.6 | 0.5 ± 0.3 | 2 ± 1 | 1.3 ± 1 | 0.5 ± 0.3 | 2.5 ± 0.5 | 1.7 ± 0.7 | 0.5 ± 0.3 | 1.2 ± 0.9 | 0.4 ± 0.3 | 0 ± 0 |
| Sum | 11 | 7 | 1.5 | 17 | 12 | 3 | 12 | 8 | 3 | 15 | 10 | 3 | 7 | 2.5 | 0 |
References
- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 1,570 View
- 86 Download
- ORCID iDs
-
Lee-Sang Hyeon

https://orcid.org/0000-0002-7608-156XDong-Kun Yang

https://orcid.org/0000-0001-5765-3043Eun-Ju Kim

https://orcid.org/0000-0003-1601-6333Yu-Ri Park

https://orcid.org/0000-0002-2829-4368Hye Jeong Lee

https://orcid.org/0000-0003-2044-6176Bang-Hun Hyun

https://orcid.org/0000-0002-3429-3425 - Related articles
-
Evaluation of serum immunoglobulin G4 concentrations in canine pancreatitis2021 March;61(1)
Clinical evaluation of a rapid diagnostic test kit for detection of canine coronavirus2018 ;58(1)
Detection of urinary trypsinogen-2 for diagnosis of canine acute pancreatitis1999 ;39(5)


 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print